March 4, 2025
Cell2fate infers RNA velocity modules to improve cell fate prediction
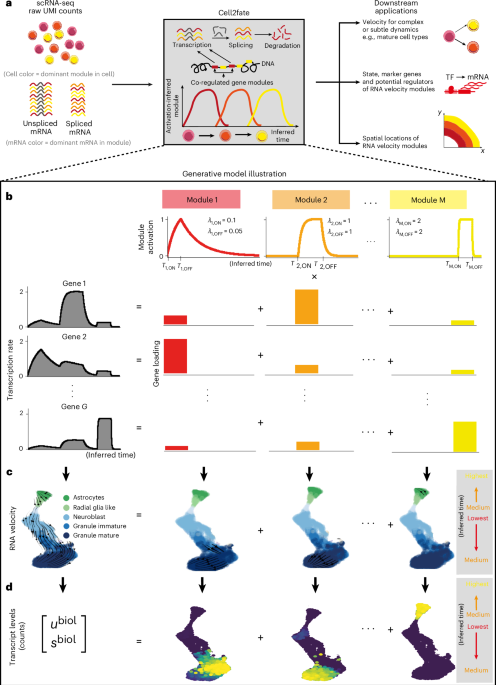
The cell2fate model Cell2fate builds on established concepts for RNA velocity1,2, employing a dynamical model to explain variation in spliced (s) and unspliced (u) read counts for individual genes and cells (Fig. 1a), which can be defined in two coupled ODEs: $$\frac{d{u}_{g}}{{dt}}={\alpha }_{g}(t)-{\beta }_{g}{u}_{g}$$ (1) $$\frac{d{s}_{g}}{{dt}}={\beta }_{g}{u}_{g}-{\gamma }_{g}{s}_{g}.$$ (2) Fig. 1: Cell2fate model overview. a–d, Cell2fate allows inferring complex and subtle transcriptional dynamics (a) by modeling gene-specific transcription rates (b) using a smaller number of independent modules with simple dynamics that also give rise to a modular structure in RNA velocity (c) and counts (d). λ denotes the rate of